Rise of the nanopores for DNA sequencing
29 Apr 2022
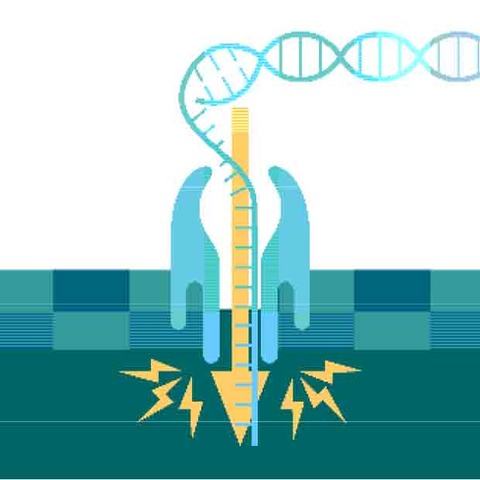
Dreamed up while driving, Deamer’s biomimetic nanopores took DNA sequencing by storm. Now, Dermot Martin follows the nanopore story from conception to an accessible method for producing solid-state membranes, with personalised medicine as one final destination
The new CBD method abolishes all of the expensive equipment required to fabricate and use solid-state nanopores.
In 1989, driving home from Eugene, Oregon, after delivering a talk, bioengineer David Deamer had a flash of inspiration – a eureka moment. He pulled over and made a rough sketch in red ink on his notebook. That sketch [1] would help change DNA mapping systems, opening a door to personalised medicine. David Deamer had ‘dreamed up’ a method for rapid nanopore DNA sequencing.
Dreaming up nanopores
It was a simple idea. If single-stranded DNA could be ‘forced’ through a tiny hole in a membrane under the influence of an electrical current, blockages due to DNA bases (A,T,C,G) would allow the reading of the entire base sequence.
At the time, nucleic acid sequencing was possible but only using slow and laborious two-dimensional chromatography. David’s sketch was a road map to a better system.
He recalls: “The idea seemed sound, yet it was two more years before I told colleague Dan Branton, who I had worked with at UC Berkeley from 1965-67. Two years after that in 1993 I visited John Kasianowitz at the US National Institute of Standards and Technology to test the idea. He knew about homopolymers of RNA and had established a way to stabilise the hemolysin pore in a lipid bilayer and also how to pass ionic current through it. We added RNA to the solution and we immediately began to see individual molecules pass through the pore, tending to block the ionic current. Quite a moment! A few months later, Dan and I repeated the experiment. We then knew that nanopore DNA sequencing might well be possible.”
A now historic paper was published in US National Academy of Sciences 1996 [2] and a patent was issued in 1998.
Biomimetic DNA sequencing
All living things are imbued with pores. They exist in the ion pumps which generate chemical energy in cells. Their role is to maintain electrolyte imbalances for nerve signalling and mediating selective biomolecular transport through cell membranes. Two particular bio-structured pores are still widely used for early research with nanopores: alpha-hemolysin (aHL), a pore derived from the toxin Staphylococcus aureus, and Mycobacterium smegmatis porin A (MspA). The earliest DNA translocation experiments were performed using aHL pores.
Today the ubiquitous MinION sequencer made by Oxford Nanopore uses a mutant of the CsgG pore, an amyloid secretion channel found in Escherichia coli, to achieve long-read sequencing. It has been a triumph and is the go-to instrument for sequencing.
Nanopipettes and solid-state nanopores
Many in the field are now turning to solid materials as an alternative to biological nanopores. Nanopipettes are thin glass capillaries extended to a nanoscale opening. Solid-state nanopores are now also being created on super-thin synthetic membranes. In practice, both these solutions operate in a very similar fashion but the longer effective lengths of nanopipettes reduce spatial sensitivity while the glass substrate lowers noise.
Unlike their biological counterparts, solid-state nanopores are robust, flexible, and can be integrated relatively easily into other solid-state electronic devices. These characteristics can make them preferable to biological pores as a long-term prospect for some applications, but there remain functionality issues linked to lower sensitivity and motion control. Advanced studies using various materials like graphene, molybdenum disulphide, silicon nitride (Si3N4), and single atomic layered phosphorene aim to address these issues.
The story of the preparation and manufacture of solid-state nanopores is fascinating. [2] In brief, planar pores are by far the types most commonly used. Until recently, planar solid-state nanopores were fabricated using transmission electron microscopes. A sub-atomic particle beam is focussed down to a nanoscale spot and a hole is drilled. The method has been operating for 20 years but it can be a long and arduous process and required persistence and dedication from practised operators. The need to apply this complex electron beam-drilling tool hindered nanopore research as only a few elite research institutions could make it work well.
Fabrication by controlled breakdown
In 2014, Northern Nanopore Instruments (NNi) introduced a controlled breakdown (CBD) method of nanopore fabrication. As described by NNi [3], CBD essentially harnesses a static shock to do the hard work. By applying voltage near the dielectric strength of the material, across an intact membrane, it is possible to drive dielectric breakdown. Essentially this is the same physical process that causes static shocks in air. Operating on the nanoscale and in the solid state, the spark will leave behind a pore on the scale of 1nm. The pore can then be tailored to any desired size in the same setup.
The new CBD method abolishes all of the expensive equipment required to fabricate and use solid-state nanopores. It has made the entire field significantly more accessible and is now a key driver of solid-state nanopore technology for rapid DNA sequencing.
Nanopore technology is evolving rapidly and is now expected to deliver faster, more accurate, and more cost-effective DNA sequencing, across a broader field of applications such as microbial resistance and pathogen surveillance in a variety of environments, from small hospitals to farms and environmental waste treatment plants. High-throughput sequencing, mapping, and comparison of cell genomes in both healthy and disease states will also drive a step-change in disease diagnosis and a paradigm shift from traditional medicine towards an era of genome-based preventive and therapeutic decisions.
Author: Dermot Martin is a freelance science writer
References
1 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6733523/

